Overrepresented sequences
On this plot you will see the number of overrepresented sequences for each sample. Tu know which sequence it is you will need to open the fastQC plot. Only sample with more than 0.1% of overrepresented sequences are represented. In blue, the plot show sequence shared by most samples. In black, the plot represent the other overrepresented sequences.
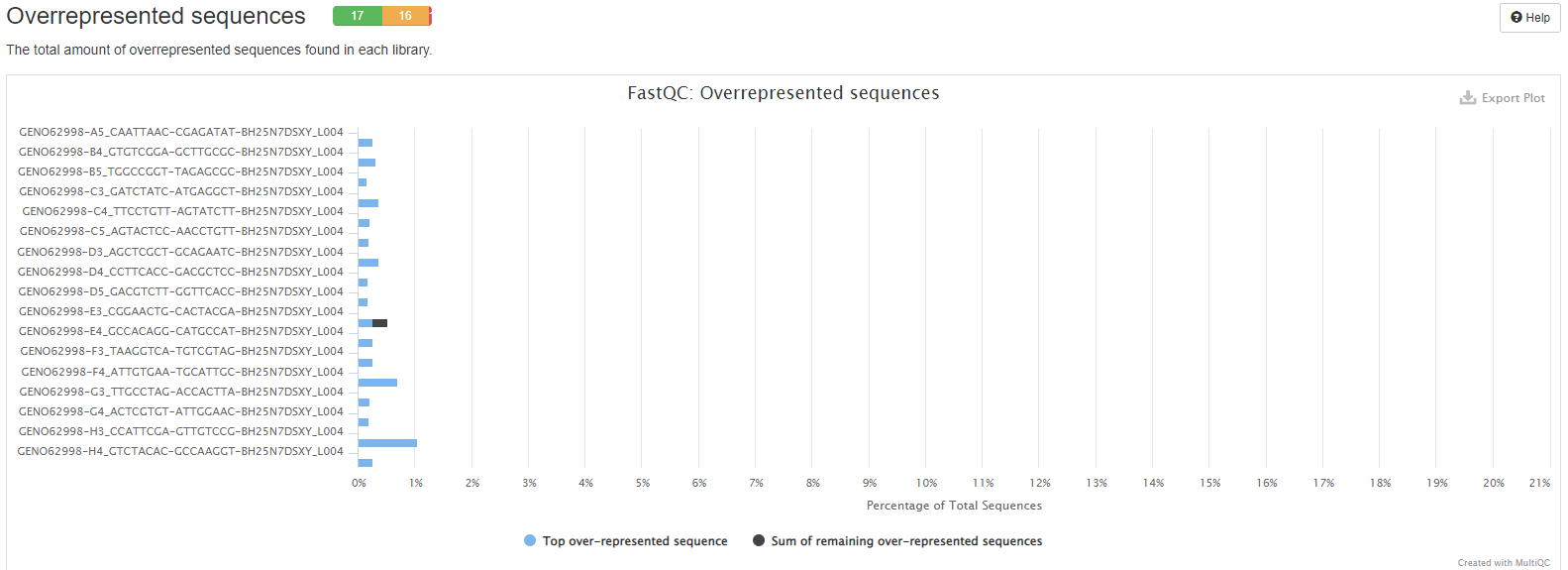
Warning
This module will issue a warning if any sequence is found to represent more than 0.1% of the total.
Failure
This module will issue an error if any sequence is found to represent more than 1% of the total.
Common reasons for warnings
This module will often be triggered when used to analyse small RNA libraries where sequences are not subjected to random fragmentation, and the same sequence may natrually be present in a significant proportion of the library.
For Novaseq sequencing if read is degradated or too short (often the case for the read 2), you will obtain here GGGGGGGGGGGGGGGGGGGGGGGGGGGGG or TTTTTTTTTTTTTTTT sequences in overrepresented sequences part in fastqQC report.
If a transcript in RNASeq is very highly expressed, you can also see a part of it in the fastQC report. In this case you can do a blast analysis to check which gene it is.