Sequence Duplication Levels
This module counts the degree of duplication for every sequence in a library and creates a plot showing the relative number of sequences with different degrees of duplication.
It identifies duplicated sequences if there is a perfect sequence match along the first 75th bases.
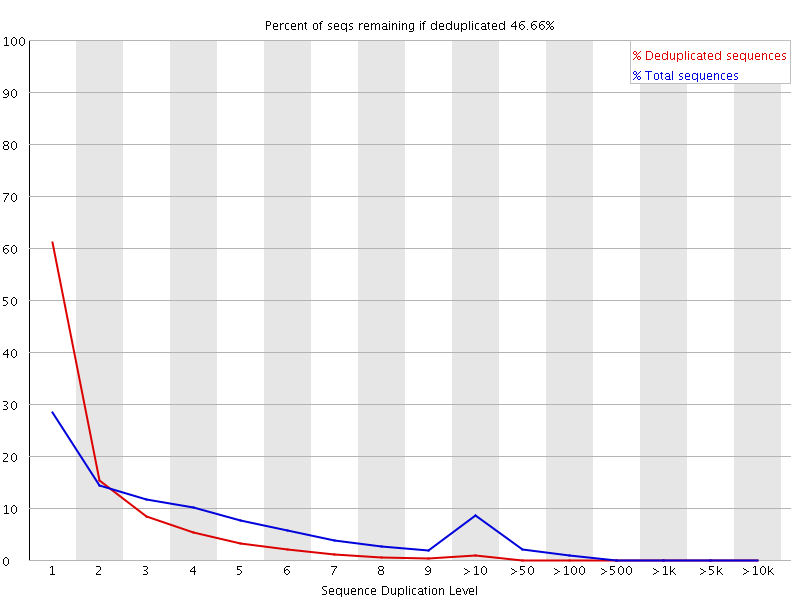
The plot shows the proportion of the library which is made up of sequences in each of the different duplication level bins. There are two lines on the plot. The blue line takes the full sequence set and shows how its duplication levels are distributed. In the red plot the sequences are de-duplicated and the proportions shown are the proportions of the deduplicated set which come from different duplication levels in the original data.
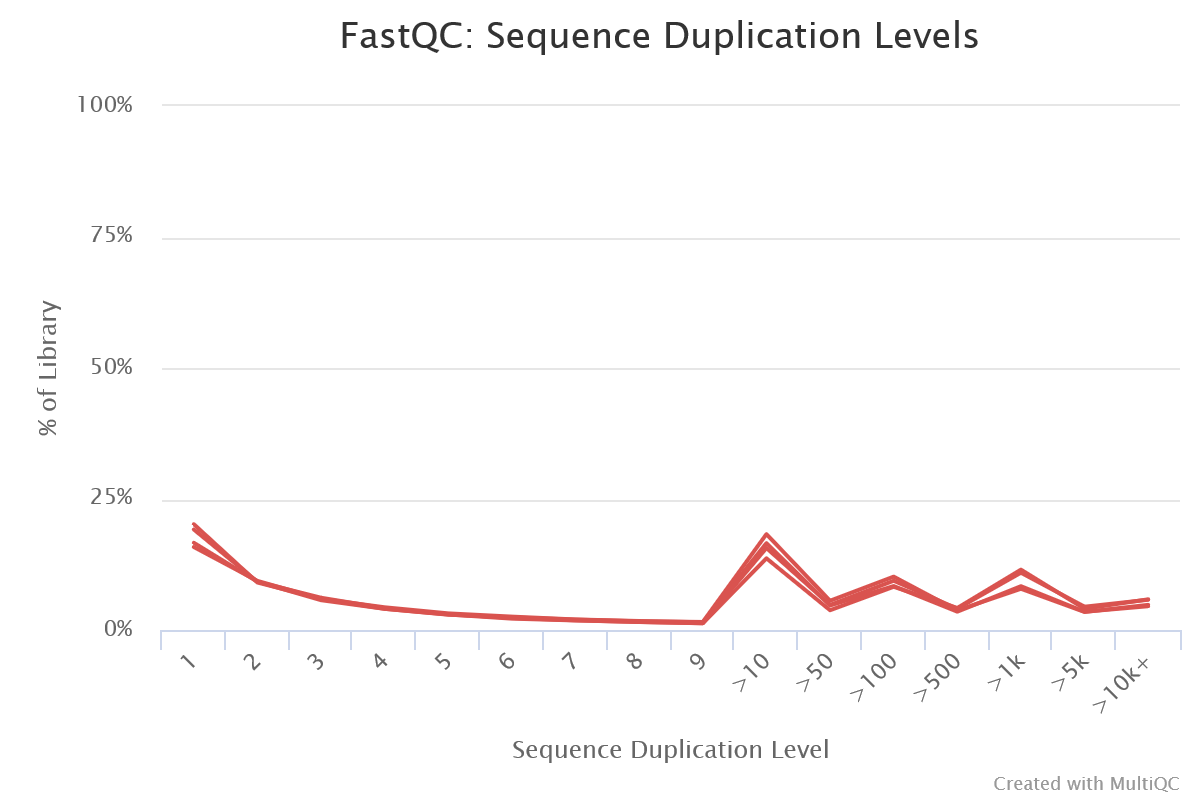
In the multiQC plot only the blue line is plotted as illusrated in next figure.
This plot can help identify a low complexity library, which could result from:
- too many cycles of PCR amplification, or
- too little starting material in quantity and/or diversity
In a diverse library most sequences will occur only once in the final set. A low level of duplication may indicate a very high level of coverage of the target sequence, but a high level of duplication is more likely to indicate some kind of enrichment bias (eg PCR over amplification).
In a properly diverse library most sequences should fall into the far left of the plot in both the red and blue lines. A general level of enrichment, indicating broad oversequencing in the library will tend to flatten the lines, lowering the low end and generally raising other categories. More specific enrichments of subsets, or the presence of low complexity contaminants will tend to produce spikes towards the right of the plot. These high duplication peaks will most often appear in the blue trace as they make up a high proportion of the original library, but usually disappear in the red trace as they make up an insignificant proportion of the deduplicated set. If peaks persist in the blue trace then this suggests that there are a large number of different highly duplicated sequences which might indicate either a contaminant set or a very severe technical duplication.
Warning
This module will issue a warning if non-unique sequences make up more than 20% of the total.
Failure
This module will issue a error if non-unique sequences make up more than 50% of the total.
Common reasons for warnings
The underlying assumption of this module is of a diverse unenriched library. Any deviation from this assumption will naturally generate duplicates and can lead to warnings or errors from this module.
In general there are two potential types of duplicate in a library, technical duplicates arising from PCR artefacts, or biological duplicates which are natural collisions where different copies of exactly the same sequence are randomly selected. From a sequence level there is no way to distinguish between these two types and both will be reported as duplicates here.
A warning or error in this module is simply a statement that you have exhausted the diversity in at least part of your library and are re-sequencing the same sequences. In a supposedly diverse library this would suggest that the diversity has been partially or completely exhausted and that you are therefore wasting sequencing capacity. However in some library types you will naturally tend to over-sequence parts of the library and therefore generate duplication and will therefore expect to see warnings or error from this module.
In RNA-Seq libraries sequences from different transcripts will be present at wildly different levels in the starting population. In order to be able to observe lowly expressed transcripts it is therefore common to greatly over-sequence high expressed transcripts, and this will potentially create large set of duplicates. This will result in high overall duplication in this test, and will often produce peaks in the higher duplication bins. This duplication will come from physically connected regions, and an examination of the distribution of duplicates in a specific genomic region will allow the distinction between over-sequencing and general technical duplication, but these distinctions are not possible from raw fastq files. A similar situation can arise in highly enriched ChIP-Seq libraries although the duplication there is less pronounced.
Finally, if you have a library where the sequence start points are constrained (a library constructed around restriction sites for example, or an unfragmented small RNA library) then the constrained start sites will generate huge dupliction levels which should not be treated as a problem, nor removed by deduplication. In these types of library you should consider using a system such as random barcoding (UMI) to allow the distinction of technical and biological duplicates.
for more informations, have a look at this post.